A recent paper by Batada and colleagues published in PLoS Biology tries to consolidate the available information on protein-protein interactions for S. cerevisiae. The authors have attempted to create a high-confidence set of interactions that they then further analyze. The main conclusion from the paper is that the highly connected proteins (usually referred to as protein hubs) do not avoid each other, as was previously put forward by other authors. From this observation they suggest that we should rethink our view of modularity in cellular networks. Cellular interaction networks should be viewed, not as altocumulus clouds, “i.e., cotton ball-like structures sparsely connected by thin wisps”, but instead as the “continuous dense aggregations of stratus clouds”.
Although I find it useful to constantly update our view of cellular networks trough the consolidation of available data, I think some words of caution remained unsaid in this work.
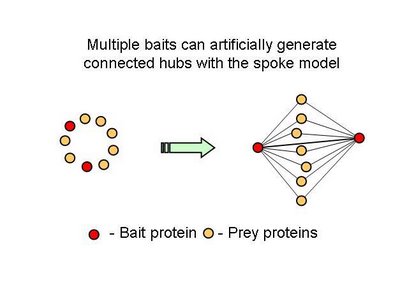
The new consolidated protein interaction network was obtained mainly from the addition of a recent curation effort from the literature, to the already available high throughput interaction datasets obtained using yeast-two-hybrid and affinity methods. The majority of the interactions added are from affinity methods. This leads me to one of points I think are not usually mentioned in this type of efforts, that not all methods will provide the same information. For example, I think that affinity methods mostly inform us that two proteins share the same complex. When a protein is tagged and used as a bait to capture prey proteins, the identified preys should belong to the same complex but it is not obvious that there should be a direct interaction between the two. However this was the assumption used here in this work. This is usually referred to as the spoke model (see figure 1).

I have tried to evaluate how likely are bait-prey interactions to occur, when compared to prey-prey interactions, using either structural information or yeast-two-hybrid interaction data (see table 1 and table 2).

Table 1 – Pull down experiments were taken from Gavin et al, 2002. For each individually reported pull down, potential bait-prey and prey-prey interactions were counted if the corresponding proteins had known PFAM domains. Using the database of domain structural interactions (iPFAM), I tried to search for plausible domain-domain interactions that could account for the protein interaction. Bait-prey interactions are roughly 2 times more likely to be explainable by a known domain-domain interactions currently stored in structural databases, than prey-prey interactions.
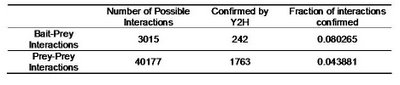 Table 2 – Pull down experiments were taken from Gavin et al, 2002. For each individually reported pull down, potential bait-prey and prey-prey interactions were counted. The overlap of these interactions with known yeast-two-hybrid interactions is shown. Bait-prey interactions are roughly 2 times more likely to be observed in a yeast-two-hybrid study than prey-prey interactions.
Table 2 – Pull down experiments were taken from Gavin et al, 2002. For each individually reported pull down, potential bait-prey and prey-prey interactions were counted. The overlap of these interactions with known yeast-two-hybrid interactions is shown. Bait-prey interactions are roughly 2 times more likely to be observed in a yeast-two-hybrid study than prey-prey interactions.What one could conclude from this is that in fact bait-prey interactions are more likely to occur than prey-prey interactions but also that a small percentage of the bait-prey interactions can be validated with a method that is more likely to measure direct interactions. Using both domain-domain structural information and yeast-two-hybrid studies, 20% of the bait-prey interactions can be accounted for. Although this value depends on our current knowledge of domain-domain interactions and the coverage of yeast-two-hybrid studied it should at least be discussed. One problem in using this model can be for example seen in figure two. When multiple baits are used for the same complex, it is easy to create artificially interacting hubs, when extrapolating binary interactions from affinity data.
In all fairness, in this study, the authors only took as a true interaction, one that was observed more than once but they also consider multiple affinity observations as confirming a direct interaction. It would be useful to come up with better methods to extrapolate from complexes to binary interactions. I hope the differences observed in this work regarding hub-hub interactions are not mainly due to the proportion of interactions extrapolated from affinity methods.
One observation that the authors used to support their claims was that, apparently, the fraction of hub-hub interactions depends on the scale of the experiment (see figure 3A taken from the authors paper). According to this result, experiments reporting higher number of interactions tend to have a lower fraction of hub-hub interactions. Hubs were defined, in the figure legend, as the 10% more connected proteins in the network, but in the results description are defined as the 5% more connected proteins. One thing that the authors failed to show is how this fraction of hub-hub interactions depends just on the size of the network. I have used the network given in the manuscript and randomly sampled 10% to 90% of the interactions (repeated 50 times) to plot the dependence of the fraction of hub-hub interactions on network size (see figure 3B). Hubs were defined as the 5% more connected proteins.

What we can see is that the fraction of hub-hub interactions depends on the size of the network, decreasing for smaller network sizes. Can this explain the result observed by the authors? In figure 3A, taken from the authors’ paper, the whole network was binned by the number of interactions reported per paper. I have tried to calculate the size of different networks obtained from binning according to the scale of the experiment (see table 3).

I would say that, although I could not reproduce exactly the same bins reported in the paper, the trend is for a decrease of network size when comparing all interactions reported in small-scale studies to those reported in medium-scale experiments. Therefore I think the observed result is mostly explained by differences in network size. In fact, if one would take all interactions reported in experiments observing at least 30 interactions the fraction of hub-hub interactions observed would be even higher than in the network obtained from very small scale experiments.
Summary
I think this manuscript highlights that we should constantly re-evaluate our views on cellular networks as more data is made available. Although the concerns raised here do not contradict their conclusions I think they should have been more carefully discussed. In particular I think we require better methods to extrapolate binary interactions from affinity methods and that it is important to mention that this might lead to false positive interactions. Also, there is a strong effect of network size on the observed fraction of hub-hub interactions. This might explain both the observed increase of hub-hub interactions with increase in the coverage and the observed dependence of the fraction of hub-hub interactions on the scale of the experiment.
Tags: