There were several interesting papers on protein interaction in the last week. For example, in PLoS Computational Biology, Kim et al described an improvement to a method that classifies interaction interfaces according to their geometry. The results of the analysis are available on their site SCOPPI.
It seems reasonable to believe that there is a limited number of protein interaction types (predicted to be around 10 thousand), much the same way that there is probably a limited number of folds used in nature. This database, along side others like the iPFAM database, provide templates on which to model other protein interactions with reasonable homology. As first proposed by Aloy and Russell, if we know that two proteins interact, for example with a yeast-two-hybrid experiment we might be able to use these databases to identity and model the interaction interface between the two proteins.
Here is an example of two complexes taken from the paper. The nodes and edges view hides the fact that RBP and RCC1 interact through different interfaces.
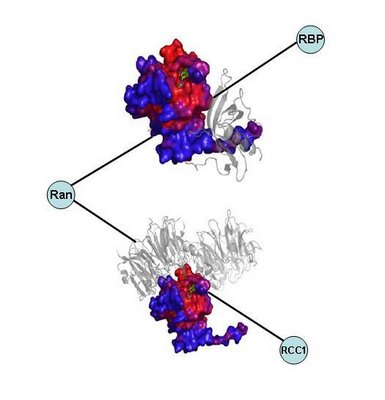
Although is has been very useful to look at protein networks as of bunch of nodes connected by edges for some analysis it would be much more informative to know what are the interacting interfaces.
The authors used the database to claim that:
- hub proteins interact with proteins using many distinct faces (what they actually show is that domains that interact with many different other domain types have more distint faces);
- two thirds of gene fusions conserve the binding orientation;
- the apparent poor conservation of interfaces is due to the diversity of interactions and partners (in my opinion it is more a suggestion that proof);
- the interfaces common to archae, bacteria and eukaryotes and mostly symmetric homo-dimers, suggesting that asymmetric and hetero interactions evolved from symmetric homo-dimers.
<speculation> Maybe if we could convert the current human interactome into more than nodes and edges we could try to see if some of disease causing polymorphisms can be explained by how they affect the interfaces. Maybe we could use this to do interaction KOs instead of whole proteins </speculation>
Tags: